Elizabeth Thrall

Assistant Professor
Physical/Biophysical Chemistry
Email: [email protected]
Office: JMH 632
Lab: JMH 628/630
Phone: 718-817-4495
-
- Assistant Professor at Fordham University
- NIH NRSA Postdoctoral Research Fellow at Harvard Medical School
- Ph.D. in Chemical Physics from Columbia University
- A.B. in Chemistry and Physics from Harvard University
-
Many biochemical processes within the cell are carried out by multi-protein complexes. I am interested in understanding how these dynamic multi-protein machines are organized, how they are remodeled to perform different functions, and how their activity is regulated to ensure that they do not act in contexts that could be harmful to the cell. Although bulk biochemistry and genetics can identify proteins involved in these processes and provide insight into mechanism, these approaches lack the molecular resolution to visualize intermediate states or distinguish between heterogeneous pathways. Using single-molecule fluorescence microscopy, I will elucidate the molecular mechanisms of the multi-protein complexes that carry out DNA replication and repair in model bacterial species.
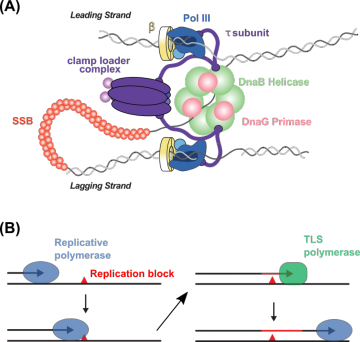
Figure 1. High and low-fidelity DNA replication. (A) Cartoon of the E. coli replisome. (B) Cartoon of high- and low-fidelity DNA polymerase exchange in translesion DNA synthesis.From the simplest forms of life to the most complex, DNA replication is performed by a multi-protein machine called the replisome. Even in a relatively simple organism, the model bacterial species Escherichia coli, the replisome is composed of 13 different proteins, most of which are present in more than one copy (Figure 1A). The replisome is held together by multiple binding interactions that span orders of magnitude in strength, and this organization appears to allow the replisome to meet a key functional challenge: being stable enough to carry out efficient DNA replication while remaining flexible enough to be remodeled when needed to carry out different tasks. My research aims to develop a molecular-level description of how the replisome and other multi-protein complexes are organized and remodeled.
New single-molecule fluorescence imaging methods are uniquely suited for these studies. These imaging techniques use genetically-encoded fluorescent tags, including fluorescent proteins (FPs) (Figure 2A). Fusing a fluorescent tag to a protein of interest allows that protein to be visualized in the cell under laser excitation in a specialized microscope. One particularly powerful approach is particle-tracking photoactivation localization microscopy (particle-tracking PALM), which takes advantages of special photoactivatable fluorescent proteins (PAFPs); these PAFPs are initially synthesized in a non-fluorescent, or dark, state, but they can be converted to a bright state by near-UV excitation. Using particle-tracking PALM, the localization, motion, and dynamics of single protein molecules can be measured in live cells (Figure 2B). As one example, the diffusion of proteins in the cell can be quantified (Figure 2C), providing information on the activity of a particular protein under conditions of interest.
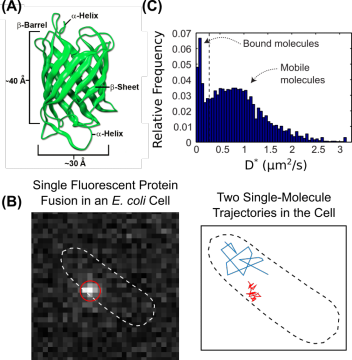
Figure 2. Single-molecule fluorescence imaging in live bacterial cells. (A) Structure of fluorescent proteins derived from Aequorea. Source: Shaner, N.C., et al. J. Cell. Sci. 2007, 120, 4227. (B) Fluorescence micrograph of a single fluorescent protein fusion in a live E. coli cell (left) and representative single-molecule trajectories in the same cell (right). (C) Distribution of apparent diffusion coefficients for the same protein in a live cell showing populations with distinct cellular mobilities.One area of research in my lab will focus on the mechanisms by which the cell switches between high-fidelity DNA replication and a lower-fidelity replication pathway called translesion synthesis (TLS). DNA is synthesized by enzymes known as DNA polymerases, but the accuracy of these enzymes can differ by many orders of magnitude. In TLS, specialized error-prone DNA polymerases are recruited to replicate DNA that has been damaged (Figure 1B). Although TLS is important for cell survival upon DNA damage, these error-prone TLS polymerases can cause mutations; for that reason, their activity must be tightly regulated in the cell. Using single-molecule fluorescence imaging, we will explore the molecular mechanisms that allow the replisome to be remodeled to allow TLS polymerases to be recruited when they are needed while excluding them during normal DNA replication.
-
Please see Google Scholar for a complete list of publications.
Thrall, E.S., Kath, J.E., Chang, S., Loparo, J.J. Single-molecule imaging reveals multiple pathways for the recruitment of translesion polymerases after DNA damage. Nature Communications, 2017, 8, 2170.
Thrall, E.S. Crowther, A.C. Yu, Z., Brus, L.E. R6G on graphene: high Raman detection sensitivity, yet decreased Raman cross-section. Nano Letters, 2012, 12, 1571.
Thrall, E.S., Preska Steinberg, A., Wu, X., Brus, L.E. The role of photon energy and semiconductor substrate in the plasmon-mediated photooxidation of citrate by silver nanoparticles. The Journal of Physical Chemistry C, 2013, 117, 26238.