The Morris Lab has previously shown that Drosophila melanogaster fried alleles disrupt CG31320 which encodes HEATR2, a HEAT Repeat protein required for cilium assembly (Diggle et al., 2014). Disruptions in the Drosophila HEATR2 gene, fried/CG31320, cause behavioral and developmental defects in larvae such as precocious wandering and early death. Before experiencing an early death, fried mutants exhibit posterior tracheal melanization as shown in figure 1. The Morris Lab used CRISPR-Cas9 and a donor template to insert the codons for a V5 tag into the endogenous fried gene. The purpose of our project is to determine the expression of Fried/HEATR2 using commercially available antibodies to detect fried-V5 in the transgenic lines of Drosophila mutant larvae. This will allow us to further analyze protein expression.
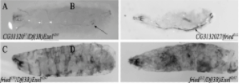
Figure 1: Drosophila fried mutants die within seven days after egg deposition following posterior tracheal melanization.
First, our Sanger Sequencing results confirmed the proper insertion of codons for a V5 tag just before the stop codon in CG31320 in two independent Drosophila lines. The expression of the fried-V5 protein in transgenic flies was then successfully confirmed using a western blot as shown in figure 3. We were able to show that fried-V5 is present in the Drosophila mutant embryos and ovaries but missing from the corresponding control groups. The western blot was also prepared for heads, wandering larvae, and non-wandering larvae. However, the western blot did not confirm the presence of fried-V5 for these. Next, we used a V5 immunostaining protocol to observe a visible representation of the CG31320/HEATR2 protein in our transgenic flies. Immunostaining was performed on first, second, and third instar larvae as well as adult heads and ovaries. The Morris Lab is still working on this experiment. However, we were able to obtain believable staining when the experiment was performed on fried embryos. After observing several slides of V5 immunoassayed transgenic embryos at different developmental stages under a Nikon fluorescence microscope, we were able to successfully show staining around the peripheral of the stage 12 transgenic embryo that was not apparent on wild type stage 12 embryos as shown in figure 2. This confirms the presence of our protein.
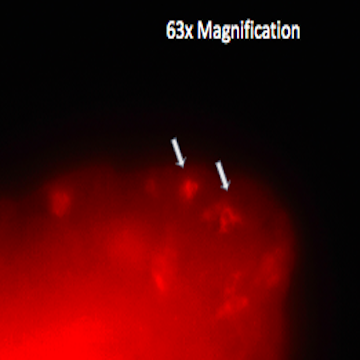
Figure 2: V5 immunostaining of a stage 12 fried-V5 transgenic embryo using TRITC. Arrows indicate staining not observed in wild type embryos.
According to our timed egg lay analysis, Df, fried150, and fried212 mutant larvae had significantly decreased viability and precociously wandered compared to their wild type counterparts. We observed that fried mutant viability declines significantly by 120 hours AED as shown in table 1. In addition, Df, fried150, and fried212 mutant larvae precociously wander as early as 72 hours AED compared to 96 hours AED in the wild type larvae as shown in table 2.
Table 1: The table shows % viability of
fried mutants. Crosses and development took place at 25C. Mutant viability declines significantly by 120 hours After Egg Deposition (AED).
| AED |
|
| 72 hours |
| |
Viability |
P |
| Df /27 |
66.67% |
P = 0.0033 |
| fried 212 /27 |
80.00% |
P = 0.0269 |
| fried 150 /27 |
58.33% |
P = 0.0076 |
|
| 96 hours |
| |
Viability |
P |
| Df /27 |
16.67% |
P = 0.2028 |
| fried 212 /27 |
8.33% |
P = 0.0093 |
| fried 150/27 |
18.18% |
P = 0.0006 |
|
| 120 hours |
| |
Viability |
P |
| Df /27 |
8.33% |
P = 0.0014 |
| fried 212 /27 |
5.97% |
P < 0.0001 |
| fried 150/27 |
13.17% |
P < 0.0001 |
|
| 144 hours |
| |
Viability |
P |
| Df /27 |
6.67% |
P = 0.0001 |
| fried 212 /27 |
6.78% |
P < 0.0001 |
| fried 150/27 |
18.75% |
P < 0.0001 |
|
| 168 hours |
| |
Viability |
P |
| Df /27 |
6.12% |
P < 0.0001 |
| fried 212 /27 |
6.33% |
P < 0.0001 |
| fried 150 /27 |
17.24% |
P < 0.0001 |
|
Table 2: The Table compares the time to wandering for fried phenotypes compared to a wild type. fried larvae precociously wander as early as 72 hours AED compared to 96 hours in wild type larvae.
| AED |
|
| 72 hours |
| |
|
Wandering (living and dead) |
Non-wandering (living) |
P* |
| Df /27 |
Wild Type |
0 |
6 |
P=0.0031 |
| |
Fried |
10 |
3 |
|
| fried 212 /27 |
Wild Type |
0 |
1 |
P=1.0000 |
| |
Fried |
2 |
2 |
|
| fried 150/27 |
Wild Type |
0 |
15 |
P=0.0307 |
| |
Fried |
7 |
17 |
|
|
| 96 hours |
| |
|
Wandering (living and dead) |
Non-wandering (living) |
P* |
| Df /27 |
Wild Type |
2 |
8 |
P=0.0001 |
| |
Fried |
12 |
0 |
|
| fried 212 /27 |
Wild Type |
5 |
17 |
P=0.0055 |
| |
Fried |
6 |
1 |
|
| fried 150/27 |
Wild Type |
0 |
90 |
P<0.0001 |
| |
Fried |
17 |
10 |
|
|
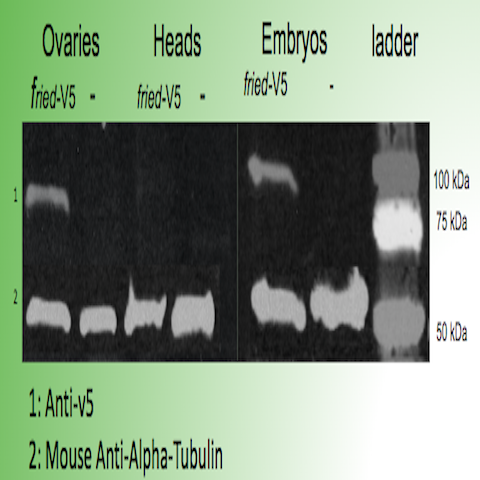
Figure 3: Western blot confirming the presence of fried-V5 in transgenic embryos and ovaries but missing from the controls.
We are currently attempting to use different Gal4 lines to express Fried/HEATR2 in all tissues or in subsets of tissues in larvae as shown by our cross-scheme in figure 4. Gal4 binds to a UAS sequence and activates transcription of the UAS-fried-V5 sequence in all of the tissues of the transgenic flies where Gal4 is expressed. We will be able to correlate the rescue of fried mutant phenotypes with expression of the transgene. So far, when using tubulin-Gal4 in which Gal4 is expressed in all tissues, all of the flies die. Therefore, we will continue to work on this rescue experiment, instead, using different Gal4 lines that will activate expression in specific tissues.

Figure 4: We can use different Gal4 lines to express Fried/HEATR2 in all tissues or in subsets of tissues in larvae.



